CDF_2010_S8591881_QCD Class Reference CDF Run II underlying event in leading jet events. More...
Detailed DescriptionCDF Run II underlying event in leading jet events. Rick Field's measurement of the underlying event in "leading jet" events. The leading jet (CDF midpoint
Definition at line 27 of file CDF_2010_S8591881_QCD.cc. Constructor & Destructor Documentation
Constructor. Definition at line 31 of file CDF_2010_S8591881_QCD.cc. : Analysis("CDF_2010_S8591881_QCD") { } Member Function Documentation
Non-templated version of string-based applyProjection, to work around header dependency issue. Definition at line 22 of file ProjectionApplier.cc. {
return evt.applyProjection(getProjection(name));
}
Non-templated version of proj-based applyProjection, to work around header dependency issue. Definition at line 28 of file ProjectionApplier.cc. {
return evt.applyProjection(proj);
}
Untemplated function to do the work... Definition at line 34 of file ProjectionApplier.cc. {
if (!_allowProjReg) {
cerr << "Trying to register projection '"
<< proj.name() << "' before init phase in '" << this->name() << "'." << endl;
exit(2);
}
const Projection& reg = getProjHandler().registerProjection(*this, proj, name);
return reg;
}
Register a data object in the histogram system.
Definition at line 795 of file Analysis.cc. {
_analysisobjects.push_back(ao);
}
Register a contained projection (user-facing version)
Definition at line 157 of file ProjectionApplier.hh. { return declareProjection(proj, name); }
List of registered analysis data objects. Definition at line 811 of file Analysis.hh. {
return _analysisobjects;
}
Analyze one event. A concrete class should here apply the necessary projections on the event and fill the relevant histograms. An overridden function must make sure it first calls the base class function. Implements Analysis. Definition at line 69 of file CDF_2010_S8591881_QCD.cc. {
/// @todo Implement Run II min bias trigger cf. CDF_2009?
const FinalState& fsj = apply<FinalState>(e, "FSJ");
if (fsj.particles().size() < 1) {
MSG_DEBUG("Failed multiplicity cut");
vetoEvent;
}
const Jets& jets = apply<FastJets>(e, "MidpointJets").jetsByPt();
MSG_DEBUG("Jet multiplicity = " << jets.size());
// We require the leading jet to be within |eta|<2
if (jets.size() < 1 || fabs(jets[0].eta()) >= 2) {
MSG_DEBUG("Failed leading jet cut");
vetoEvent;
}
const double jetphi = jets[0].phi();
const double jeteta = jets[0].eta();
const double jetpT = jets[0].pT();
MSG_DEBUG("Leading jet: pT = " << jetpT
<< ", eta = " << jeteta << ", phi = " << jetphi);
// Get the event weight
const double weight = e.weight();
// Get the final states to work with for filling the distributions
const FinalState& cfs = apply<ChargedFinalState>(e, "CFS");
size_t numOverall(0), numToward(0), numAway(0) ;
long int numTrans1(0), numTrans2(0);
double ptSumOverall(0.0), ptSumToward(0.0), ptSumTrans1(0.0), ptSumTrans2(0.0), ptSumAway(0.0);
double ptMaxOverall(0.0), ptMaxToward(0.0), ptMaxTrans1(0.0), ptMaxTrans2(0.0), ptMaxAway(0.0);
// Calculate all the charged stuff
foreach (const Particle& p, cfs.particles()) {
const double dPhi = deltaPhi(p.phi(), jetphi);
const double pT = p.pT();
const double phi = p.phi();
double rotatedphi = phi - jetphi;
while (rotatedphi < 0) rotatedphi += 2*PI;
ptSumOverall += pT;
++numOverall;
if (pT > ptMaxOverall) {
ptMaxOverall = pT;
}
if (dPhi < PI/3.0) {
ptSumToward += pT;
++numToward;
if (pT > ptMaxToward) ptMaxToward = pT;
}
else if (dPhi < 2*PI/3.0) {
if (rotatedphi <= PI) {
ptSumTrans1 += pT;
++numTrans1;
if (pT > ptMaxTrans1) ptMaxTrans1 = pT;
} else {
ptSumTrans2 += pT;
++numTrans2;
if (pT > ptMaxTrans2) ptMaxTrans2 = pT;
}
}
else {
ptSumAway += pT;
++numAway;
if (pT > ptMaxAway) ptMaxAway = pT;
}
} // end charged particle loop
// Fill the histograms
_hist_tnchg->fill(jetpT/GeV, numToward/(4*PI/3), weight);
_hist_pnchg->fill(jetpT/GeV, (numTrans1+numTrans2)/(4*PI/3), weight);
_hist_pmaxnchg->fill(jetpT/GeV, (numTrans1>numTrans2 ? numTrans1 : numTrans2)/(2*PI/3), weight);
_hist_pminnchg->fill(jetpT/GeV, (numTrans1<numTrans2 ? numTrans1 : numTrans2)/(2*PI/3), weight);
_hist_pdifnchg->fill(jetpT/GeV, abs(numTrans1-numTrans2)/(2*PI/3), weight);
_hist_anchg->fill(jetpT/GeV, numAway/(4*PI/3), weight);
_hist_tcptsum->fill(jetpT/GeV, ptSumToward/GeV/(4*PI/3), weight);
_hist_pcptsum->fill(jetpT/GeV, (ptSumTrans1+ptSumTrans2)/GeV/(4*PI/3), weight);
_hist_pmaxcptsum->fill(jetpT/GeV, (ptSumTrans1>ptSumTrans2 ? ptSumTrans1 : ptSumTrans2)/GeV/(2*PI/3), weight);
_hist_pmincptsum->fill(jetpT/GeV, (ptSumTrans1<ptSumTrans2 ? ptSumTrans1 : ptSumTrans2)/GeV/(2*PI/3), weight);
_hist_pdifcptsum->fill(jetpT/GeV, fabs(ptSumTrans1-ptSumTrans2)/GeV/(2*PI/3), weight);
_hist_acptsum->fill(jetpT/GeV, ptSumAway/GeV/(4*PI/3), weight);
if ((numTrans1+numTrans2) > 0) {
_hist_pcptave->fill(jetpT/GeV, (ptSumTrans1+ptSumTrans2)/GeV/(numTrans1+numTrans2), weight);
_hist_pcptmax->fill(jetpT/GeV, (ptMaxTrans1 > ptMaxTrans2 ? ptMaxTrans1 : ptMaxTrans2)/GeV, weight);
}
}
Apply the supplied projection on event evt (user-facing alias).
Definition at line 80 of file ProjectionApplier.hh. { return applyProjection<PROJ>(evt, proj); }
Apply the supplied projection on event evt (user-facing alias).
Definition at line 92 of file ProjectionApplier.hh. { return applyProjection<PROJ>(evt, proj); }
Apply the supplied projection on event evt (user-facing alias).
Definition at line 104 of file ProjectionApplier.hh. { return applyProjection<PROJ>(evt, name); }
Apply the supplied projection on event evt.
Definition at line 74 of file ProjectionApplier.hh. {
return pcast<PROJ>(_applyProjection(evt, proj));
}
Apply the supplied projection on event evt.
Definition at line 86 of file ProjectionApplier.hh. {
return pcast<PROJ>(_applyProjection(evt, proj));
}
Apply the named projection on event evt.
Definition at line 98 of file ProjectionApplier.hh. {
return pcast<PROJ>(_applyProjection(evt, name));
}
Helper for histogram asymmetry calculation.
Definition at line 674 of file Analysis.cc. {
const string path = s->path();
*s = YODA::asymm(*h1, *h2);
s->setPath(path);
}
Helper for histogram asymmetry calculation.
Names & emails of paper/analysis authors. Names and email of authors in 'NAME <EMAIL>' format. The first name in the list should be the primary contact person. Definition at line 133 of file Analysis.hh. Incoming beam IDs for this run. Definition at line 33 of file Analysis.cc.
Incoming beams for this run. Definition at line 29 of file Analysis.cc.
BibTeX citation key for this article. Definition at line 186 of file Analysis.hh.
BibTeX citation entry for this article. Definition at line 191 of file Analysis.hh.
Book a counter. Definition at line 196 of file Analysis.cc. {
// const string& xtitle,
// const string& ytitle) {
const string path = histoPath(cname);
CounterPtr ctr = make_shared<Counter>(path, title);
addAnalysisObject(ctr);
MSG_TRACE("Made counter " << cname << " for " << name());
// hist->setAnnotation("XLabel", xtitle);
// hist->setAnnotation("YLabel", ytitle);
return ctr;
}
Book a counter, using a path generated from the dataset and axis ID codes The paper, dataset and x/y-axis IDs will be used to build the histo name in the HepData standard way. Definition at line 210 of file Analysis.cc. {
// const string& xtitle,
// const string& ytitle) {
const string axisCode = makeAxisCode(datasetId, xAxisId, yAxisId);
return bookCounter(axisCode, title);
}
Book a 1D histogram with nbins uniformly distributed across the range lower - upper . Definition at line 219 of file Analysis.cc. {
const string path = histoPath(hname);
Histo1DPtr hist = make_shared<Histo1D>(nbins, lower, upper, path, title);
addAnalysisObject(hist);
MSG_TRACE("Made histogram " << hname << " for " << name());
hist->setAnnotation("XLabel", xtitle);
hist->setAnnotation("YLabel", ytitle);
return hist;
}
Book a 1D histogram with non-uniform bins defined by the vector of bin edges binedges . Definition at line 234 of file Analysis.cc. {
const string path = histoPath(hname);
Histo1DPtr hist = make_shared<Histo1D>(binedges, path, title);
addAnalysisObject(hist);
MSG_TRACE("Made histogram " << hname << " for " << name());
hist->setAnnotation("XLabel", xtitle);
hist->setAnnotation("YLabel", ytitle);
return hist;
}
Book a 1D histogram with binning from a reference scatter. Definition at line 249 of file Analysis.cc. {
const string path = histoPath(hname);
Histo1DPtr hist = make_shared<Histo1D>(refscatter, path);
addAnalysisObject(hist);
MSG_TRACE("Made histogram " << hname << " for " << name());
hist->setTitle(title);
hist->setAnnotation("XLabel", xtitle);
hist->setAnnotation("YLabel", ytitle);
return hist;
}
Book a 1D histogram, using the binnings in the reference data histogram. Definition at line 265 of file Analysis.cc. {
const Scatter2D& refdata = refData(hname);
return bookHisto1D(hname, refdata, title, xtitle, ytitle);
}
Book a 1D histogram, using the binnings in the reference data histogram. The paper, dataset and x/y-axis IDs will be used to build the histo name in the HepData standard way. Definition at line 274 of file Analysis.cc. {
const string axisCode = makeAxisCode(datasetId, xAxisId, yAxisId);
return bookHisto1D(axisCode, title, xtitle, ytitle);
}
Book a 2D histogram with nxbins and nybins uniformly distributed across the ranges xlower - xupper and ylower - yupper respectively along the x- and y-axis.
Definition at line 289 of file Analysis.cc. {
const string path = histoPath(hname);
Histo2DPtr hist = make_shared<Histo2D>(nxbins, xlower, xupper, nybins, ylower, yupper, path, title);
addAnalysisObject(hist);
MSG_TRACE("Made 2D histogram " << hname << " for " << name());
hist->setAnnotation("XLabel", xtitle);
hist->setAnnotation("YLabel", ytitle);
hist->setAnnotation("ZLabel", ztitle);
return hist;
}
Book a 2D histogram with non-uniform bins defined by the vectorx of bin edges xbinedges and ybinedges. Definition at line 308 of file Analysis.cc. {
const string path = histoPath(hname);
Histo2DPtr hist = make_shared<Histo2D>(xbinedges, ybinedges, path, title);
addAnalysisObject(hist);
MSG_TRACE("Made 2D histogram " << hname << " for " << name());
hist->setAnnotation("XLabel", xtitle);
hist->setAnnotation("YLabel", ytitle);
hist->setAnnotation("ZLabel", ztitle);
return hist;
}
Book a 1D profile histogram with nbins uniformly distributed across the range lower - upper . Definition at line 368 of file Analysis.cc. {
const string path = histoPath(hname);
Profile1DPtr prof = make_shared<Profile1D>(nbins, lower, upper, path, title);
addAnalysisObject(prof);
MSG_TRACE("Made profile histogram " << hname << " for " << name());
prof->setAnnotation("XLabel", xtitle);
prof->setAnnotation("YLabel", ytitle);
return prof;
}
Book a 1D profile histogram with non-uniform bins defined by the vector of bin edges binedges . Definition at line 383 of file Analysis.cc. {
const string path = histoPath(hname);
Profile1DPtr prof = make_shared<Profile1D>(binedges, path, title);
addAnalysisObject(prof);
MSG_TRACE("Made profile histogram " << hname << " for " << name());
prof->setAnnotation("XLabel", xtitle);
prof->setAnnotation("YLabel", ytitle);
return prof;
}
Book a 1D profile histogram with binning from a reference scatter. Definition at line 398 of file Analysis.cc. {
const string path = histoPath(hname);
Profile1DPtr prof = make_shared<Profile1D>(refscatter, path);
addAnalysisObject(prof);
MSG_TRACE("Made profile histogram " << hname << " for " << name());
prof->setTitle(title);
prof->setAnnotation("XLabel", xtitle);
prof->setAnnotation("YLabel", ytitle);
return prof;
}
Book a 1D profile histogram, using the binnings in the reference data histogram. Definition at line 414 of file Analysis.cc. {
const Scatter2D& refdata = refData(hname);
return bookProfile1D(hname, refdata, title, xtitle, ytitle);
}
Book a 1D profile histogram, using the binnings in the reference data histogram. The paper, dataset and x/y-axis IDs will be used to build the histo name in the HepData standard way. Definition at line 423 of file Analysis.cc. {
const string axisCode = makeAxisCode(datasetId, xAxisId, yAxisId);
return bookProfile1D(axisCode, title, xtitle, ytitle);
}
Book a 2D profile histogram with nxbins and nybins uniformly distributed across the ranges xlower - xupper and ylower - yupper respectively along the x- and y-axis. Definition at line 436 of file Analysis.cc. {
const string path = histoPath(hname);
Profile2DPtr prof = make_shared<Profile2D>(nxbins, xlower, xupper, nybins, ylower, yupper, path, title);
addAnalysisObject(prof);
MSG_TRACE("Made 2D profile histogram " << hname << " for " << name());
prof->setAnnotation("XLabel", xtitle);
prof->setAnnotation("YLabel", ytitle);
prof->setAnnotation("ZLabel", ztitle);
return prof;
}
Book a 2D profile histogram with non-uniform bins defined by the vectorx of bin edges xbinedges and ybinedges. Definition at line 455 of file Analysis.cc. {
const string path = histoPath(hname);
Profile2DPtr prof = make_shared<Profile2D>(xbinedges, ybinedges, path, title);
addAnalysisObject(prof);
MSG_TRACE("Made 2D profile histogram " << hname << " for " << name());
prof->setAnnotation("XLabel", xtitle);
prof->setAnnotation("YLabel", ytitle);
prof->setAnnotation("ZLabel", ztitle);
return prof;
}
Book a 2-dimensional data point set with the given name.
Definition at line 525 of file Analysis.cc. {
Scatter2DPtr s;
const string path = histoPath(hname);
if (copy_pts) {
const Scatter2D& refdata = refData(hname);
s = make_shared<Scatter2D>(refdata, path);
foreach (Point2D& p, s->points()) p.setY(0, 0);
} else {
s = make_shared<Scatter2D>(path);
}
addAnalysisObject(s);
MSG_TRACE("Made scatter " << hname << " for " << name());
s->setTitle(title);
s->setAnnotation("XLabel", xtitle);
s->setAnnotation("YLabel", ytitle);
return s;
}
Book a 2-dimensional data point set, using the binnings in the reference data histogram. The paper, dataset and x/y-axis IDs will be used to build the histo name in the HepData standard way.
Definition at line 515 of file Analysis.cc. {
const string axisCode = makeAxisCode(datasetId, xAxisId, yAxisId);
return bookScatter2D(axisCode, copy_pts, title, xtitle, ytitle);
}
Book a 2-dimensional data point set with equally spaced x-points in a range. The y values and errors will be set to 0. Definition at line 548 of file Analysis.cc. {
const string path = histoPath(hname);
Scatter2DPtr s = make_shared<Scatter2D>(path);
const double binwidth = (upper-lower)/npts;
for (size_t pt = 0; pt < npts; ++pt) {
const double bincentre = lower + (pt + 0.5) * binwidth;
s->addPoint(bincentre, 0, binwidth/2.0, 0);
}
addAnalysisObject(s);
MSG_TRACE("Made scatter " << hname << " for " << name());
s->setTitle(title);
s->setAnnotation("XLabel", xtitle);
s->setAnnotation("YLabel", ytitle);
return s;
}
Book a 2-dimensional data point set based on provided contiguous "bin edges". The y values and errors will be set to 0. Definition at line 569 of file Analysis.cc. {
const string path = histoPath(hname);
Scatter2DPtr s = make_shared<Scatter2D>(path);
for (size_t pt = 0; pt < binedges.size()-1; ++pt) {
const double bincentre = (binedges[pt] + binedges[pt+1]) / 2.0;
const double binwidth = binedges[pt+1] - binedges[pt];
s->addPoint(bincentre, 0, binwidth/2.0, 0);
}
addAnalysisObject(s);
MSG_TRACE("Made scatter " << hname << " for " << name());
s->setTitle(title);
s->setAnnotation("XLabel", xtitle);
s->setAnnotation("YLabel", ytitle);
return s;
}
Collider on which the experiment ran. Definition at line 171 of file Analysis.hh.
Get the process cross-section in pb. Throws if this hasn't been set. Definition at line 151 of file Analysis.cc. {
if (!_gotCrossSection || std::isnan(_crossSection)) {
string errMsg = "You did not set the cross section for the analysis " + name();
throw Error(errMsg);
}
return _crossSection;
}
Get the process cross-section per generated event in pb. Throws if this hasn't been set. Definition at line 159 of file Analysis.cc. {
const double sumW = sumOfWeights();
assert(sumW != 0.0);
return _crossSection / sumW;
}
Register a contained projection (user-facing version)
Definition at line 151 of file ProjectionApplier.hh. { return declareProjection(proj, name); }
Register a contained projection. The type of the argument is used to instantiate a new projection internally: this new object is applied to events rather than the argument object. Hence you are advised to only use locally-scoped Projection objects in your Projection and Analysis constructors, and to avoid polymorphism (e.g. handling
Definition at line 142 of file ProjectionApplier.hh. {
const Projection& reg = _declareProjection(proj, name);
const PROJ& rtn = dynamic_cast<const PROJ&>(reg);
return rtn;
}
Get a full description of the analysis. Full textual description of this analysis, what it is useful for, what experimental techniques are applied, etc. Should be treated as a chunk of restructuredText (http://docutils.sourceforge.net/rst.html), with equations to be rendered as LaTeX with amsmath operators. Definition at line 152 of file Analysis.hh. {
return info().description();
}
Helper for counter division.
Definition at line 593 of file Analysis.cc.
Helper for histogram division with raw YODA objects.
Helper for histogram division.
Definition at line 606 of file Analysis.cc.
Helper for histogram division with raw YODA objects.
Helper for profile histogram division.
Definition at line 619 of file Analysis.cc.
Helper for profile histogram division with raw YODA objects.
Helper for 2D histogram division.
Definition at line 632 of file Analysis.cc.
Helper for 2D histogram division with raw YODA objects.
Helper for 2D profile histogram division.
Definition at line 645 of file Analysis.cc.
Helper for 2D profile histogram division with raw YODA objects
Helper for histogram efficiency calculation.
Definition at line 661 of file Analysis.cc. {
const string path = s->path();
*s = YODA::efficiency(*h1, *h2);
s->setPath(path);
}
Helper for histogram efficiency calculation.
Experiment which performed and published this analysis. Definition at line 166 of file Analysis.hh. {
return info().experiment();
}
Finalize this analysis object. A concrete class should here make all necessary operations on the histograms. Writing the histograms to a file is, however, done by the Rivet class. An overridden function must make sure it first calls the base class function. Reimplemented from Analysis. Definition at line 163 of file CDF_2010_S8591881_QCD.cc. {
}
Get the named projection, specifying return type via a template argument (user-facing alias).
Definition at line 57 of file ProjectionApplier.hh. { return getProjection<PROJ>(name); }
Get a data object from the histogram system
Definition at line 828 of file Analysis.hh. {
foreach (const AnalysisObjectPtr& ao, analysisObjects()) {
if (ao->path() == histoPath(name)) return dynamic_pointer_cast<AO>(ao);
}
throw Exception("Data object " + histoPath(name) + " not found");
}
Get a data object from the histogram system (non-const)
Definition at line 839 of file Analysis.hh. {
foreach (const AnalysisObjectPtr& ao, analysisObjects()) {
if (ao->path() == histoPath(name)) return dynamic_pointer_cast<AO>(ao);
}
throw Exception("Data object " + histoPath(name) + " not found");
}
Get a named Histo1D object from the histogram system. Definition at line 854 of file Analysis.hh. {
return getAnalysisObject<Histo1D>(name);
}
Get a named Histo1D object from the histogram system (non-const) Definition at line 859 of file Analysis.hh. {
return getAnalysisObject<Histo1D>(name);
}
Get a Histo1D object from the histogram system by axis ID codes (non-const) Definition at line 864 of file Analysis.hh. {
return getAnalysisObject<Histo1D>(makeAxisCode(datasetId, xAxisId, yAxisId));
}
Get a Histo1D object from the histogram system by axis ID codes (non-const) Definition at line 869 of file Analysis.hh. {
return getAnalysisObject<Histo1D>(makeAxisCode(datasetId, xAxisId, yAxisId));
}
Get a Log object based on the name() property of the calling analysis object. Reimplemented from ProjectionApplier. Definition at line 78 of file Analysis.cc. {
string logname = "Rivet.Analysis." + name();
return Log::getLog(logname);
}
Get a named Profile1D object from the histogram system. Definition at line 896 of file Analysis.hh. {
return getAnalysisObject<Profile1D>(name);
}
Get a named Profile1D object from the histogram system (non-const) Definition at line 901 of file Analysis.hh. {
return getAnalysisObject<Profile1D>(name);
}
Get a Profile1D object from the histogram system by axis ID codes (non-const) Definition at line 906 of file Analysis.hh. {
return getAnalysisObject<Profile1D>(makeAxisCode(datasetId, xAxisId, yAxisId));
}
Get a Profile1D object from the histogram system by axis ID codes (non-const) Definition at line 911 of file Analysis.hh. {
return getAnalysisObject<Profile1D>(makeAxisCode(datasetId, xAxisId, yAxisId));
}
Get the named projection, specifying return type via a template argument.
Definition at line 50 of file ProjectionApplier.hh. {
const Projection& p = getProjHandler().getProjection(*this, name);
return pcast<PROJ>(p);
}
Get the named projection (non-templated, so returns as a reference to a Projection base class). Definition at line 61 of file ProjectionApplier.hh. {
return getProjHandler().getProjection(*this, name);
}
Get the contained projections, including recursion. Definition at line 43 of file ProjectionApplier.hh. {
return getProjHandler().getChildProjections(*this, ProjectionHandler::DEEP);
}
Get a reference to the ProjectionHandler for this thread. Definition at line 122 of file ProjectionApplier.hh. {
return _projhandler;
}
Get a named Scatter2D object from the histogram system. Definition at line 938 of file Analysis.hh. {
return getAnalysisObject<Scatter2D>(name);
}
Get a named Scatter2D object from the histogram system (non-const) Definition at line 943 of file Analysis.hh. {
return getAnalysisObject<Scatter2D>(name);
}
Get a Scatter2D object from the histogram system by axis ID codes (non-const) Definition at line 948 of file Analysis.hh. {
return getAnalysisObject<Scatter2D>(makeAxisCode(datasetId, xAxisId, yAxisId));
}
Get a Scatter2D object from the histogram system by axis ID codes (non-const) Definition at line 953 of file Analysis.hh. {
return getAnalysisObject<Scatter2D>(makeAxisCode(datasetId, xAxisId, yAxisId));
}
Access the controlling AnalysisHandler object. Definition at line 289 of file Analysis.hh. { return *_analysishandler; }
Get the canonical histogram "directory" path for this analysis.
Definition at line 38 of file Analysis.cc. {
/// @todo Cache in a member variable
string _histoDir;
if (_histoDir.empty()) {
_histoDir = "/" + name();
if (handler().runName().length() > 0) {
_histoDir = "/" + handler().runName() + _histoDir;
}
replace_all(_histoDir, "//", "/"); //< iterates until none
}
return _histoDir;
}
Get the canonical histogram path for the named histogram in this analysis. Definition at line 52 of file Analysis.cc. {
const string path = histoDir() + "/" + hname;
return path;
}
Get the canonical histogram path for the numbered histogram in this analysis. Definition at line 58 of file Analysis.cc. {
return histoDir() + "/" + makeAxisCode(datasetId, xAxisId, yAxisId);
}
Get the actual AnalysisInfo object in which all this metadata is stored. Definition at line 105 of file Analysis.hh.
Get the actual AnalysisInfo object in which all this metadata is stored (non-const). Definition at line 247 of file Analysis.hh.
Initialize this analysis object. A concrete class should here book all necessary histograms. An overridden function must make sure it first calls the base class function. Reimplemented from Analysis. Definition at line 40 of file CDF_2010_S8591881_QCD.cc. {
// Final state for the jet finding
const FinalState fsj(-4.0, 4.0, 0.0*GeV);
declare(fsj, "FSJ");
declare(FastJets(fsj, FastJets::CDFMIDPOINT, 0.7), "MidpointJets");
// Charged final state for the distributions
const ChargedFinalState cfs(-1.0, 1.0, 0.5*GeV);
declare(cfs, "CFS");
// Book histograms
_hist_tnchg = bookProfile1D(10, 1, 1);
_hist_pnchg = bookProfile1D(10, 1, 2);
_hist_anchg = bookProfile1D(10, 1, 3);
_hist_pmaxnchg = bookProfile1D(11, 1, 1);
_hist_pminnchg = bookProfile1D(11, 1, 2);
_hist_pdifnchg = bookProfile1D(11, 1, 3);
_hist_tcptsum = bookProfile1D(12, 1, 1);
_hist_pcptsum = bookProfile1D(12, 1, 2);
_hist_acptsum = bookProfile1D(12, 1, 3);
_hist_pmaxcptsum = bookProfile1D(13, 1, 1);
_hist_pmincptsum = bookProfile1D(13, 1, 2);
_hist_pdifcptsum = bookProfile1D(13, 1, 3);
_hist_pcptave = bookProfile1D(14, 1, 1);
_hist_pcptmax = bookProfile1D(15, 1, 1);
}
Get the Inspire ID code for this analysis. Definition at line 120 of file Analysis.hh.
Helper for converting a differential histo to an integral one.
Definition at line 774 of file Analysis.cc.
Helper for converting a differential histo to an integral one.
Definition at line 781 of file Analysis.cc.
Check if analysis is compatible with the provided beam particle IDs and energies. Definition at line 97 of file Analysis.cc. {
return isCompatible(beams.first.pid(), beams.second.pid(),
beams.first.energy(), beams.second.energy());
}
Check if analysis is compatible with the provided beam particle IDs and energies. Definition at line 103 of file Analysis.cc. {
PdgIdPair beams(beam1, beam2);
pair<double,double> energies(e1, e2);
return isCompatible(beams, energies);
}
Check if analysis is compatible with the provided beam particle IDs and energies.
Get the internal histogram name for given d, x and y (cf. HepData) Definition at line 63 of file Analysis.cc. {
stringstream axisCode;
axisCode << "d";
if (datasetId < 10) axisCode << 0;
axisCode << datasetId;
axisCode << "-x";
if (xAxisId < 10) axisCode << 0;
axisCode << xAxisId;
axisCode << "-y";
if (yAxisId < 10) axisCode << 0;
axisCode << yAxisId;
return axisCode.str();
}
Mark object as owned by the _projhandler
Definition at line 111 of file ProjectionApplier.hh. { _owned = true; }
Get the name of the analysis. By default this is computed by combining the results of the experiment, year and Spires ID metadata methods and you should only override it if there's a good reason why those won't work. Implements ProjectionApplier. Definition at line 115 of file Analysis.hh. {
return (info().name().empty()) ? _defaultname : info().name();
}
Return true if this analysis needs to know the process cross-section.
Definition at line 230 of file Analysis.hh. {
return info().needsCrossSection();
}
Normalize the given histogram, histo, to area = norm. Definition at line 706 of file Analysis.cc. {
if (!histo) {
MSG_WARNING("Failed to normalize histo=NULL in analysis " << name() << " (norm=" << norm << ")");
return;
}
MSG_TRACE("Normalizing histo " << histo->path() << " to " << norm);
try {
histo->normalize(norm, includeoverflows);
} catch (YODA::Exception& we) {
MSG_WARNING("Could not normalize histo " << histo->path());
return;
}
}
Normalize the given histograms, histos, to area = norm.
Definition at line 662 of file Analysis.hh. {
for (auto& h : histos) normalize(h, norm, includeoverflows);
}
Definition at line 667 of file Analysis.hh. {
for (auto& h : histos) normalize(h, norm, includeoverflows);
}
Normalize the given histogram, histo, to area = norm. Definition at line 740 of file Analysis.cc. {
if (!histo) {
MSG_ERROR("Failed to normalize histo=NULL in analysis " << name() << " (norm=" << norm << ")");
return;
}
MSG_TRACE("Normalizing histo " << histo->path() << " to " << norm);
try {
histo->normalize(norm, includeoverflows);
} catch (YODA::Exception& we) {
MSG_WARNING("Could not normalize histo " << histo->path());
return;
}
}
Normalize the given histograms, histos, to area = norm.
Definition at line 693 of file Analysis.hh. {
for (auto& h : histos) normalize(h, norm, includeoverflows);
}
Definition at line 698 of file Analysis.hh. {
for (auto& h : histos) normalize(h, norm, includeoverflows);
}
Get the number of events seen (via the analysis handler). Use in the finalize phase only. Definition at line 84 of file Analysis.cc.
Get reference data for a named histo
Definition at line 179 of file Analysis.cc.
Get reference data for a numbered histo
Definition at line 190 of file Analysis.cc. {
const string hname = makeAxisCode(datasetId, xAxisId, yAxisId);
return refData(hname);
}
Get reference data for a named histo
Definition at line 350 of file Analysis.hh.
Get reference data for a numbered histo
Definition at line 366 of file Analysis.hh. {
const string hname = makeAxisCode(datasetId, xAxisId, yAxisId);
return refData(hname);
}
Journal, and preprint references. Definition at line 181 of file Analysis.hh. {
return info().references();
}
Unregister a data object from the histogram system (by name) Definition at line 799 of file Analysis.cc. {
for (vector<AnalysisObjectPtr>::iterator it = _analysisobjects.begin(); it != _analysisobjects.end(); ++it) {
if ((*it)->path() == path) {
_analysisobjects.erase(it);
break;
}
}
}
Unregister a data object from the histogram system (by pointer) Definition at line 808 of file Analysis.cc. {
for (vector<AnalysisObjectPtr>::iterator it = _analysisobjects.begin(); it != _analysisobjects.end(); ++it) {
if (*it == ao) {
_analysisobjects.erase(it);
break;
}
}
}
Return the allowed pairs of incoming beams required by this analysis. Definition at line 207 of file Analysis.hh.
Sets of valid beam energy pairs, in GeV. Definition at line 218 of file Analysis.hh.
Information about the events needed as input for this analysis. Event types, energies, kinematic cuts, particles to be considered stable, etc. etc. Should be treated as a restructuredText bullet list (http://docutils.sourceforge.net/rst.html) Definition at line 161 of file Analysis.hh.
Multiplicatively scale the given counter, cnt, by factor factor. Definition at line 687 of file Analysis.cc. {
if (!cnt) {
MSG_WARNING("Failed to scale counter=NULL in analysis " << name() << " (scale=" << factor << ")");
return;
}
if (std::isnan(factor) || std::isinf(factor)) {
MSG_WARNING("Failed to scale counter=" << cnt->path() << " in analysis: " << name() << " (invalid scale factor = " << factor << ")");
factor = 0;
}
MSG_TRACE("Scaling counter " << cnt->path() << " by factor " << factor);
try {
cnt->scaleW(factor);
} catch (YODA::Exception& we) {
MSG_WARNING("Could not scale counter " << cnt->path());
return;
}
}
Multiplicatively scale the given counters, cnts, by factor factor.
Definition at line 645 of file Analysis.hh. {
for (auto& c : cnts) scale(c, factor);
}
Definition at line 650 of file Analysis.hh. {
// for (size_t i = 0; i < std::extent<decltype(cnts)>::value; ++i) scale(cnts[i], factor);
for (auto& c : cnts) scale(c, factor);
}
Multiplicatively scale the given histogram, histo, by factor factor. Definition at line 721 of file Analysis.cc. {
if (!histo) {
MSG_WARNING("Failed to scale histo=NULL in analysis " << name() << " (scale=" << factor << ")");
return;
}
if (std::isnan(factor) || std::isinf(factor)) {
MSG_WARNING("Failed to scale histo=" << histo->path() << " in analysis: " << name() << " (invalid scale factor = " << factor << ")");
factor = 0;
}
MSG_TRACE("Scaling histo " << histo->path() << " by factor " << factor);
try {
histo->scaleW(factor);
} catch (YODA::Exception& we) {
MSG_WARNING("Could not scale histo " << histo->path());
return;
}
}
Multiplicatively scale the given histograms, histos, by factor factor.
Definition at line 677 of file Analysis.hh. {
for (auto& h : histos) scale(h, factor);
}
Multiplicatively scale the given histogram, histo, by factor factor. Definition at line 755 of file Analysis.cc. {
if (!histo) {
MSG_ERROR("Failed to scale histo=NULL in analysis " << name() << " (scale=" << factor << ")");
return;
}
if (std::isnan(factor) || std::isinf(factor)) {
MSG_ERROR("Failed to scale histo=" << histo->path() << " in analysis: " << name() << " (invalid scale factor = " << factor << ")");
factor = 0;
}
MSG_TRACE("Scaling histo " << histo->path() << " by factor " << factor);
try {
histo->scaleW(factor);
} catch (YODA::Exception& we) {
MSG_WARNING("Could not scale histo " << histo->path());
return;
}
}
Multiplicatively scale the given histograms, histos, by factor factor.
Definition at line 708 of file Analysis.hh. {
for (auto& h : histos) scale(h, factor);
}
Set the cross section from the generator. Definition at line 145 of file Analysis.cc. {
_crossSection = xs;
_gotCrossSection = true;
return *this;
}
Declare whether this analysis needs to know the process cross-section from the generator.
Definition at line 235 of file Analysis.hh. {
info().setNeedsCrossSection(needed);
return *this;
}
Declare the allowed pairs of incoming beams required by this analysis. Definition at line 211 of file Analysis.hh.
Declare the list of valid beam energy pairs, in GeV. Definition at line 222 of file Analysis.hh. {
info().setEnergies(requiredEnergies);
return *this;
}
Get the SPIRES ID code for this analysis (~deprecated). Definition at line 125 of file Analysis.hh.
Centre of mass energy for this run. Definition at line 25 of file Analysis.cc.
Get a short description of the analysis. Short (one sentence) description used as an index entry. Use description() to provide full descriptive paragraphs of analysis details. Definition at line 142 of file Analysis.hh.
Get the sum of event weights seen (via the analysis handler). Use in the finalize phase only. Definition at line 89 of file Analysis.cc. {
return handler().sumOfWeights();
}
Any work to be done on this analysis. Definition at line 201 of file Analysis.hh.
When the original experimental analysis was published. Definition at line 176 of file Analysis.hh. Member Data Documentation
Flag to forbid projection registration in analyses until the init phase. Definition at line 176 of file ProjectionApplier.hh.
Definition at line 179 of file CDF_2010_S8591881_QCD.cc.
Definition at line 173 of file CDF_2010_S8591881_QCD.cc.
Definition at line 183 of file CDF_2010_S8591881_QCD.cc.
Definition at line 184 of file CDF_2010_S8591881_QCD.cc.
Definition at line 178 of file CDF_2010_S8591881_QCD.cc.
Definition at line 182 of file CDF_2010_S8591881_QCD.cc.
Definition at line 176 of file CDF_2010_S8591881_QCD.cc.
Definition at line 180 of file CDF_2010_S8591881_QCD.cc.
Definition at line 174 of file CDF_2010_S8591881_QCD.cc.
Definition at line 181 of file CDF_2010_S8591881_QCD.cc.
Definition at line 175 of file CDF_2010_S8591881_QCD.cc.
Definition at line 172 of file CDF_2010_S8591881_QCD.cc.
Definition at line 177 of file CDF_2010_S8591881_QCD.cc.
Definition at line 171 of file CDF_2010_S8591881_QCD.cc. The documentation for this class was generated from the following file: Generated on Tue Dec 13 2016 16:33:36 for The Rivet MC analysis system by  1.7.6.1
1.7.6.1
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
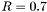 ) must be within
) must be within  and defines the "toward" phi direction. Particles are selected in
and defines the "toward" phi direction. Particles are selected in 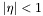 . For the
. For the 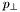 -related observables there is a
-related observables there is a 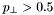 GeV cut. For
GeV cut. For 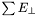 there is no
there is no 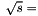 1960 GeV
1960 GeV  cutoffs are probably required to fill the profile histograms:
cutoffs are probably required to fill the profile histograms:  0 (min bias), 10, 20, 50, 100, 150 GeV
0 (min bias), 10, 20, 50, 100, 150 GeV 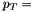 0, 30, 50, 80, 130, 180 GeV
0, 30, 50, 80, 130, 180 GeV